GENOMIC
& transcriptomiC
The platform offers genome and transcriptome analysis tools, as well as expertise for NGS projects (RNAseq, Single cell and Spatial omics…)
TEAM
Expertises
EQUIPMENTS
BOOKING/CONTACT
TEAM

Emeline LHUILLIER

Rachel Fourdin
SCIENTIFIC COMMITTEE
| Anaïs BRIOT | Email me |
| Jean-Philippe PRADERE | Email me |
| Emmanuelle ARNAUD | Email me |
| Aline MAIRAL | Email me |
| Alexandra MONTAGNER | Email me |
| Eric LACAZETTE | Email me |
| Gaëtan CHICANNE | Email me |
| Audrey CASEMAYOU | Email me |
| Adeline CHAUBET | Email me |
| Constandina ARVANITIS | Email me |
| Carine VALLE | Email me |
| Alexia ZAKAROFF-GIRARD | Email me |
| Sabrina BENAOUADI | Email me |
| Thibaut DUPARC | Email me |
| Nicole MALET | Email me |
EXPERTISES
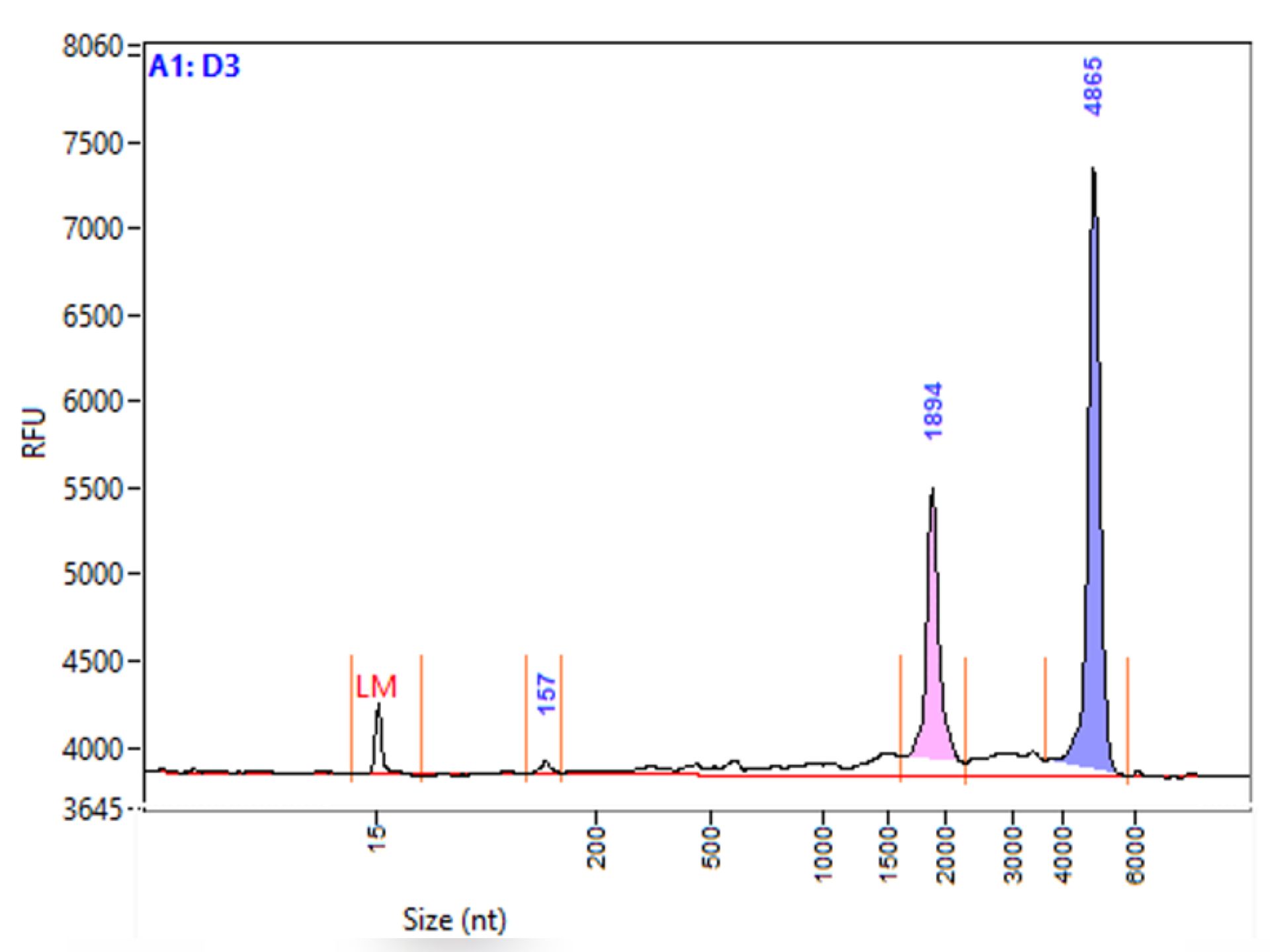
DNA/RNA QUALITY CONTROL
Provision of instruments (Nanodrop, Fragment Analyzer, etc.), training in their use or performance of controls, assistance in data interpretation.
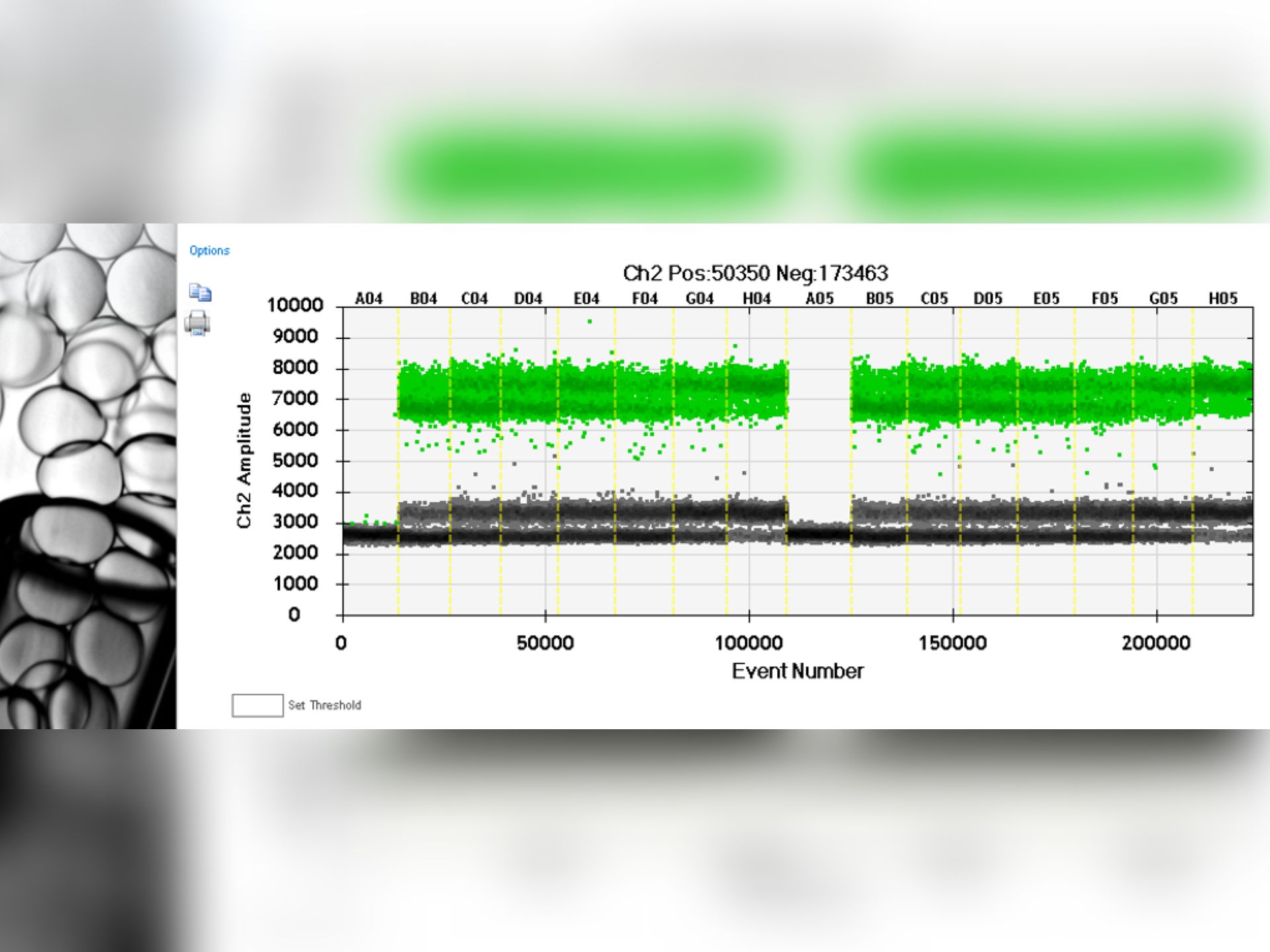
QPCR / DDPCR
Provision of several instruments: qPCR 96 or 384 wells, QX200.
Training in their use and support in the design of projects according to the applications.
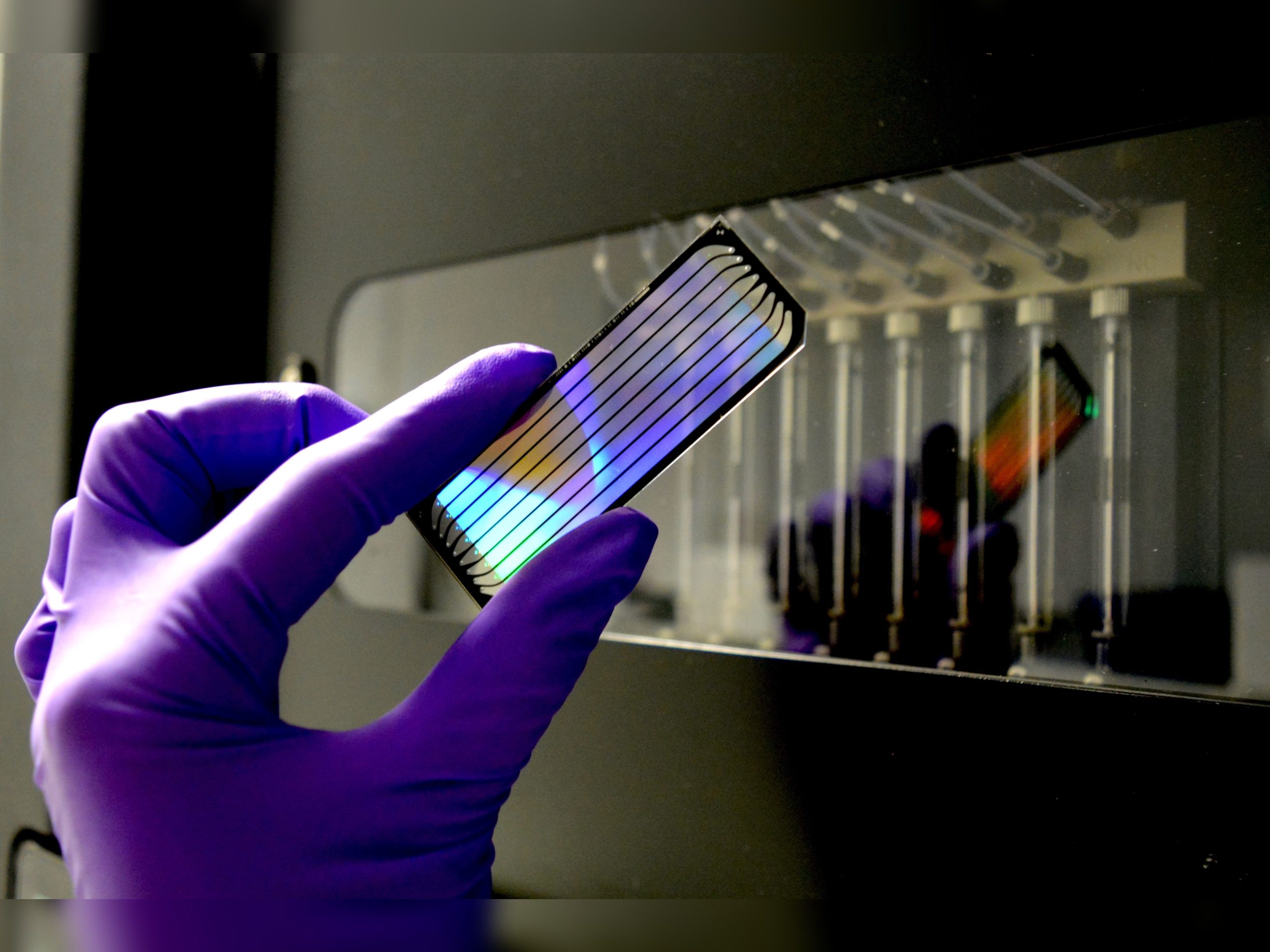
NGS LIBRARY DEVELOPMENT
RNAseq, ChIPseq, WGS…: support in project design, library construction, coordination with sequencing and bioinfo analysis platforms.
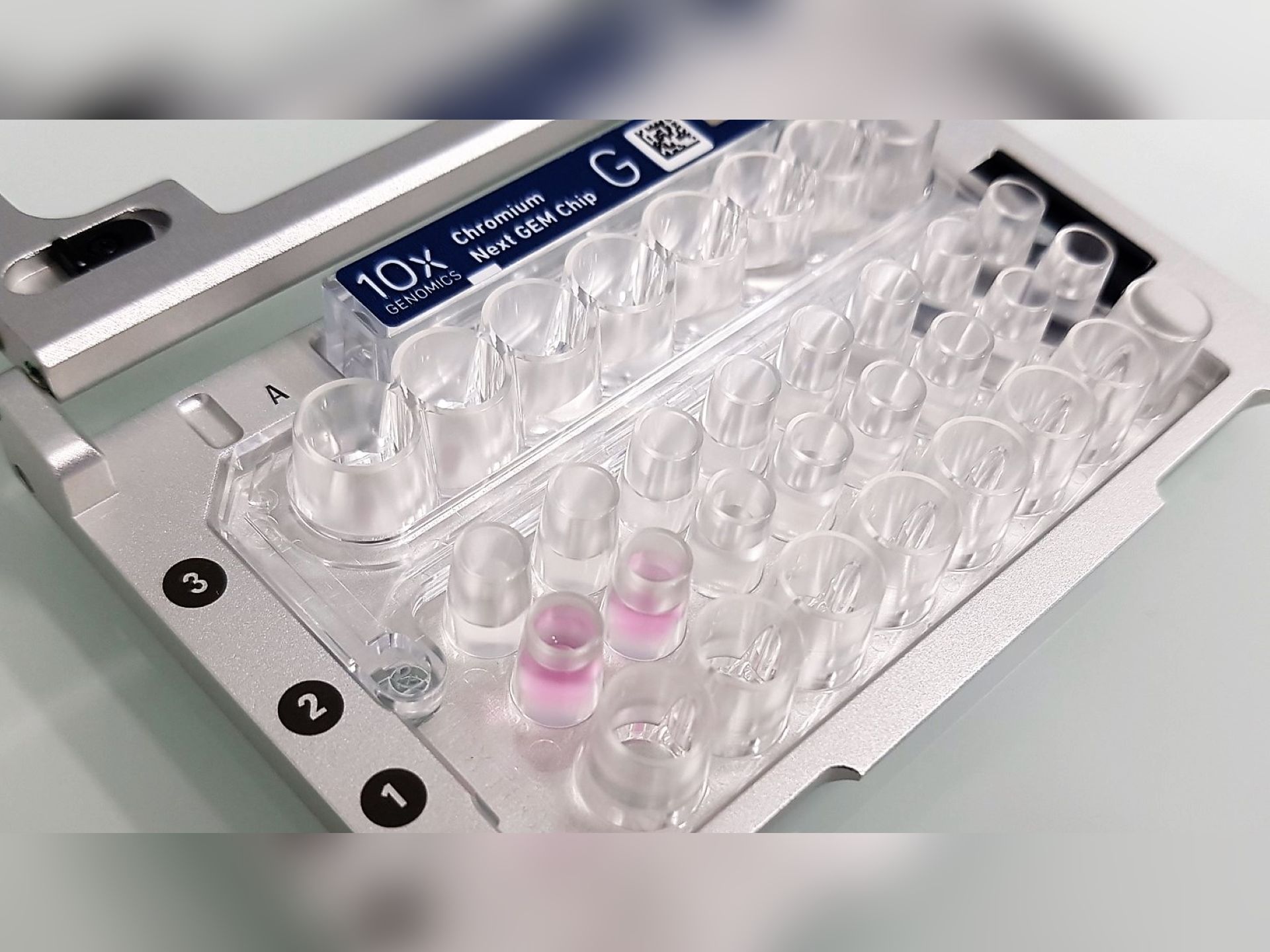
SINGLE CELL & SPATIALS OMICS
Taking charge of projects in close interaction with the research team (collaboration, partnership) of single cell 3′ RNAseq, CITE-seq, 5’VDJ, cell hashing, etc…
EQUIPMENTS
Here is an exhaustive list of the equipment offered by the GeT-Santé platform…)
BOOKING & CONTACT
Booking schedule for instruments that can be used independently after training and accreditation
A request ?
Actualités
Symposium du PEPR MED-OOC
Vendredi 6 février, I2MC Le programme de...
Winter Scientific Symposium de l’ISHR
Ne manquez pas le Winter Scientific Symposium de...
Dominique Langin sur France 3 Occitanie
Dominique Langin will be the guest on the...
RECENT publications
Tamoxifen Accelerates Endothelial Healing by Targeting ERα in Smooth Muscle Cell. Zahreddine R, Davezac M, Smirnova NF, Buscato M, Lhuillier E, Lupieri A, Solinhac R, Vinel A, Vessieres E, Henrion D, Renault M-A, Gadeau A-P, Flouriot G, Lenfant F, Laffargue M, Metivier R, Arnal J-F, Fontaine C.
Circulation Research, 2020. Pubmed
Single-cell RNA sequencing unveils the shared and the distinct cytotoxic hallmarks of human TCRVδ1 and TCRVδ2 γδ T lymphocytes. Pizzolato G, Kaminski H, Tosolini M, Franchini D-M, Pont F, Martins F, Valle C, Labourdette D, Cadot S, Quillet-Mary A, Poupot M, Laurent C, Ysebaert L, Meraviglia S, Dieli F, Merville P, Milpied P, Déchanet-Merville J, Fournié J-J. Proceedings of the National Academy of Sciences, 2019. Pubmed
Single-Cell Analysis Reveals Heterogeneity of High Endothelial Venules and Different Regulation of Genes Controlling Lymphocyte Entry to Lymph Nodes. Veerman K, Tardiveau C, Martins F, Coudert J, Girard J-P. Cell Reports, 2019. Pubmed
Niacin induces miR-502-3p expression which impairs insulin sensitivity in human adipocytes. Montastier E, Beuzelin D, Martins F, Mir L, Marqués MA, Thalamas C, Iacovoni J, Langin D, Viguerie N. Int J Obes (Lond). 2018. Pubmed
Caloric Restriction and Diet-Induced Weight Loss Do Not Induce Browning of Human Subcutaneous White Adipose Tissue in Women and Men with Obesity. Barquissau V, Léger B, Beuzelin D, Martins F, Amri EZ, Pisani DF, Saris WHM, Astrup A, Maoret JJ, Iacovoni J, Déjean S, Moro C, Viguerie N, Langin D. Cell Rep. 2018. Pubmed


Inserm/UPS UMR 1297 - I2MC Institut des Maladies Métaboliques et Cardiovasculaires
1 avenue Jean Poulhès - BP 84225 - 31432 Toulouse Cedex 4
Tél. : 05 61 32 56 00
Horaires
Du lundi au vendredi
8h30 - 12h30 / 13h45 -16h45








